Abstract
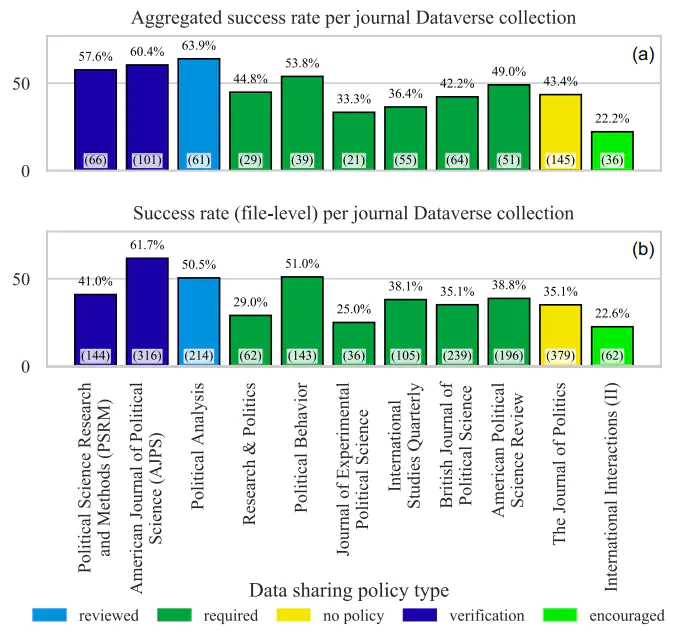
This article presents a study on the quality and execution of research code from publicly-available replication datasets at the Harvard Dataverse repository. Research code is typically created by a group of scientists and published together with academic papers to facilitate research transparency and reproducibility. For this study, we define ten questions to address aspects impacting research reproducibility and reuse. First, we retrieve and analyze more than 2000 replication datasets with over 9000 unique R files published from 2010 to 2020. Second, we execute the code in a clean runtime environment to assess its ease of reuse. Common coding errors were identified, and some of them were solved with automatic code cleaning to aid code execution. We find that 74% of R files failed to complete without error in the initial execution, while 56% failed when code cleaning was applied, showing that many errors can be prevented with good coding practices. We also analyze the replication datasets from journals’ collections and discuss the impact of the journal policy strictness on the code re-execution rate. Finally, based on our results, we propose a set of recommendations for code dissemination aimed at researchers, journals, and repositories.